Motivation
The rapid development of Next Generation Sequencing (NGS) technologies is significantly lowering the cost of human genome data sequencing and making the genomic information readily available for a variety medical applications, especially precise medicine for individuals with specific genetic conditions. The exponentially growing volume of genome data generated is becoming a bottleneck for wide adoption of Genome data guided precision medicine applications. Current Genome data file formats like FastQ and SAM/BAM offer very limited compression efficiency due to very shallow modeling of the underlying generative models that create the DNA sequences, for example, typical human genome data with approx. 200 reads coverage and quality score, is approx. 1.5TB in FastQ format, while the aligned data representation and compression with BAM still requires approx. 300GB. This is a prohibitive cost in communication and storage for the everyday precision medicine applications.
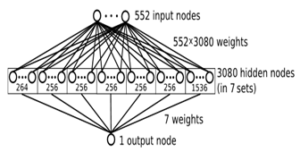
Shallow Neural Network Like Context Modeling for High Efficiency Arithmetic Coding (click image for link to Dr. Li’s site)
Objectives
In this work, we will partner with Tsinghua University/Flora Production to develop novel compression schemes for un-aligned and aligned Genome sequence data, capitalizing on the recent development in deep learning tools, especially the RNN (Recurrent Neural Network) tools that can derive a better DNA sequence context and prediction model, which in turn is mated with a hierarchical-context arithmetic coding scheme, to achieve at least 100% gains in compression efficiency. Our initial work with a modified shallow neural network based on PAQ codec is already showing 40% compression efficiency gains over the current state of art. For the online compression part, the parallel high throughput on-line compression exploiting the mobile device GPU architecture, significantly reducing the compression delay, is also crucial for practical applications. How to design context merge and skip modes to accelerate the arithmetic coding in addition to the parallel approach can deliver even more gains in compression speed.
Deliverables
The first year deliverables will be focusing on the modeling part, with RNN based solution for better arithmetic coding context models and prediction, The second year will focus on the mobile device GPU based implementation of the online Genome compression algorithms, with high throughput low encoding delay objectives. Acceleration comes in forms of parallel processing with GPU, context merge and skipping, faster RNN model evaluation with model compression and hardware acceleration. A proper streaming data format will also be developed to support over the Internet low delay communication and random access.
Experimental Plan
The new Genome data compression and streaming solution will be deployed to our research partner, Children’s Mercy Hospital (CMH) in Kansas City, to test various scenarios. Compression efficiency, low delay and high throughput end-to-end delivery will be the key test.
Principal Investigator
Zhu Li, Associate Professor – Department of Computer Science and Electrical Engineering, University of Missouri, Kansas City
Acknowledgements
We wish to thank NSF, Samsung Research America, Snapchat, and Qualcomm.
